2.3. Uncertainties in the theoretical predictions#
In this tutorial, you will learn how to use the microscopic module to print out some microscopic predictions for thermodynamical properties.
Import the libraries that will be employed in this tutorial.
# Import numpy
import numpy as np
# Import matplotlib
import matplotlib.pyplot as plt
# Import nucleardatapy package
import nucleardatapy as nuda
You can simply print out the properties of the nuda’s function that we will use:
# Explore the nucleardatapy module to find the correct attribute
print(dir(nuda.matter.setupMicro))
['__class__', '__delattr__', '__dict__', '__dir__', '__doc__', '__eq__', '__format__', '__ge__', '__getattribute__', '__getstate__', '__gt__', '__hash__', '__init__', '__init_subclass__', '__le__', '__lt__', '__module__', '__ne__', '__new__', '__reduce__', '__reduce_ex__', '__repr__', '__setattr__', '__sizeof__', '__str__', '__subclasshook__', '__weakref__', 'init_self', 'print_outputs']
Get the full list of models:
models, models_lower = nuda.matter.micro_models()
print('models:',models)
models: ['1981-VAR-AM-FP', '1998-VAR-AM-APR', '1998-VAR-AM-APR-fit', '2012-AFDMC-NM-RES-1', '2012-AFDMC-NM-RES-2', '2012-AFDMC-NM-RES-3', '2012-AFDMC-NM-RES-4', '2012-AFDMC-NM-RES-5', '2012-AFDMC-NM-RES-6', '2012-AFDMC-NM-RES-7', '2012-AFDMC-NM-FIT-1', '2012-AFDMC-NM-FIT-2', '2012-AFDMC-NM-FIT-3', '2012-AFDMC-NM-FIT-4', '2012-AFDMC-NM-FIT-5', '2012-AFDMC-NM-FIT-6', '2012-AFDMC-NM-FIT-7', '2022-AFDMC-NM', '2024-BHF-AM-2BF-Av18', '2024-BHF-AM-2BF-BONN', '2024-BHF-AM-2BF-CDBONN', '2024-BHF-AM-2BF-NSC97a', '2024-BHF-AM-2BF-NSC97b', '2024-BHF-AM-2BF-NSC97c', '2024-BHF-AM-2BF-NSC97d', '2024-BHF-AM-2BF-NSC97e', '2024-BHF-AM-2BF-NSC97f', '2006-BHF-AM-Av18', '2024-BHF-AM-23BF-Av18', '2024-BHF-AM-23BF-BONN', '2024-BHF-AM-23BF-CDBONN', '2024-BHF-AM-23BF-NSC97a', '2024-BHF-AM-23BF-NSC97b', '2024-BHF-AM-23BF-NSC97c', '2024-BHF-AM-23BF-NSC97d', '2024-BHF-AM-23BF-NSC97e', '2024-BHF-AM-23BF-NSC97f', '2008-QMC-NM-swave', '2010-QMC-NM-AV4', '2009-DLQMC-NM', '2014-AFQMC-NM', '2016-QMC-NM', '2018-QMC-NM', '2024-QMC-NM', '2013-MBPT-NM', '2016-MBPT-AM', '2019-MBPT-AM-L59', '2019-MBPT-AM-L69', '2020-MBPT-AM', '2024-NLEFT-AM']
Fix one model for which we want to extract the data:
model = '2013-QMC-NM'
mic = nuda.matter.setupMicro( model = model )
model -> 2013-QMC-NM
setup_micro: The model name 2013-QMC-NM is not in the list of models.
setup_micro: list of models: ['1981-VAR-AM-FP', '1998-VAR-AM-APR', '1998-VAR-AM-APR-fit', '2012-AFDMC-NM-RES-1', '2012-AFDMC-NM-RES-2', '2012-AFDMC-NM-RES-3', '2012-AFDMC-NM-RES-4', '2012-AFDMC-NM-RES-5', '2012-AFDMC-NM-RES-6', '2012-AFDMC-NM-RES-7', '2012-AFDMC-NM-FIT-1', '2012-AFDMC-NM-FIT-2', '2012-AFDMC-NM-FIT-3', '2012-AFDMC-NM-FIT-4', '2012-AFDMC-NM-FIT-5', '2012-AFDMC-NM-FIT-6', '2012-AFDMC-NM-FIT-7', '2022-AFDMC-NM', '2024-BHF-AM-2BF-Av18', '2024-BHF-AM-2BF-BONN', '2024-BHF-AM-2BF-CDBONN', '2024-BHF-AM-2BF-NSC97a', '2024-BHF-AM-2BF-NSC97b', '2024-BHF-AM-2BF-NSC97c', '2024-BHF-AM-2BF-NSC97d', '2024-BHF-AM-2BF-NSC97e', '2024-BHF-AM-2BF-NSC97f', '2006-BHF-AM-Av18', '2024-BHF-AM-23BF-Av18', '2024-BHF-AM-23BF-BONN', '2024-BHF-AM-23BF-CDBONN', '2024-BHF-AM-23BF-NSC97a', '2024-BHF-AM-23BF-NSC97b', '2024-BHF-AM-23BF-NSC97c', '2024-BHF-AM-23BF-NSC97d', '2024-BHF-AM-23BF-NSC97e', '2024-BHF-AM-23BF-NSC97f', '2008-QMC-NM-swave', '2010-QMC-NM-AV4', '2009-DLQMC-NM', '2014-AFQMC-NM', '2016-QMC-NM', '2018-QMC-NM', '2024-QMC-NM', '2013-MBPT-NM', '2016-MBPT-AM', '2019-MBPT-AM-L59', '2019-MBPT-AM-L69', '2020-MBPT-AM', '2024-NLEFT-AM']
setup_micro: -- Exit the code --
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[4], line 2
1 model = '2013-QMC-NM'
----> 2 mic = nuda.matter.setupMicro( model = model )
File ~/work/nucleardatapy/nucleardatapy/version-1.0/nucleardatapy/matter/setup_micro.py:380, in setupMicro.__init__(self, model, var1, var2)
378 print("setup_micro: list of models:", models)
379 print("setup_micro: -- Exit the code --")
--> 380 exit()
381 #
382 # ==============================
383 # Read files associated to model
384 # ==============================
385 #
386 self.nm_rmass = nuda.cst.mnc2
NameError: name 'exit' is not defined
print the outputs:
mic.print_outputs()
- Print output:
model: 2013-QMC-NM
ref: I. Tews et al., PRL 110, 032504 (2013)
label: QMC-2013
note: write here notes about this EOS.
self.sm_den: None
self.sm_effmass: None
den: [0.01 0.031 0.051 0.072 0.092 0.113 0.133 0.154 0.174 0.195 0.215 0.236
0.256 0.277 0.297 0.318 0.338 0.359 0.379 0.4 ] in fm$^{-3}$
kfn: [0.01 0.031 0.051 0.072 0.092 0.113 0.133 0.154 0.174 0.195 0.215 0.236
0.256 0.277 0.297 0.318 0.338 0.359 0.379 0.4 ] in fm$^{-1}$
asy: 0.0
nm_den: [0.04 0.06 0.08 0.1 0.12 0.14 0.16 0.18 0.2 ] in fm$^{-3}$
nm_kfn: [1.058 1.211 1.333 1.436 1.526 1.606 1.68 1.747 1.809] in fm$^{-1}$
nm_chempot: [ 9.172 12.018 15.248 19.158 23.751 29.183 35.202 42.038 49.605] in MeV
nm_e2a: [ 5.934 7.342 8.779 10.467 12.356 14.559 16.783 19.225 21.766] in MeV
nm_e2a_err: [0.556 1.043 1.495 1.912 2.372 2.829 3.337 3.82 4.241] in MeV
nm_e2v: [0.237 0.44 0.702 1.047 1.483 2.038 2.685 3.46 4.353] in MeV fm$^{-3}$
nm_e2v_err: [0.022 0.063 0.12 0.191 0.285 0.396 0.534 0.688 0.848] in MeV fm$^{-3}$
nm_pre: [0.13 0.281 0.518 0.869 1.367 2.047 2.947 4.106 5.568] in MeV fm$^{-3}$
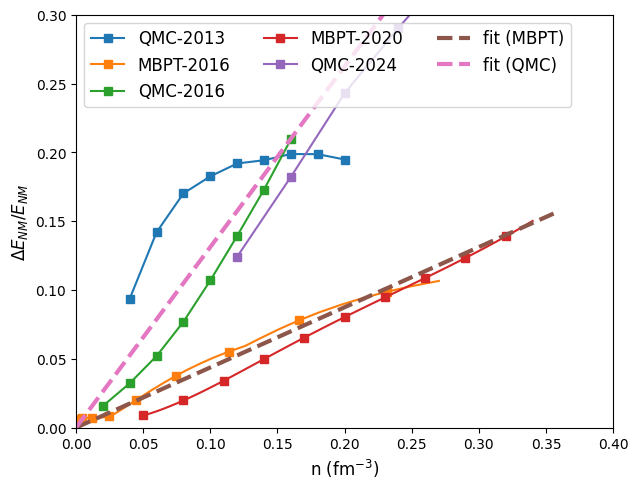
The uncertianty is defined as:
print('uncertainties:',mic.nm_e2a_err/mic.nm_e2a)
uncertainties: [0.09374429 0.14202996 0.17025439 0.18262481 0.19196782 0.19433464
0.1988345 0.19871935 0.19484428]
Fix a list of models instead of a unique one
models = [ '2013-QMC-NM', '2016-MBPT-AM', '2016-QMC-NM', '2020-MBPT-AM', '2024-QMC-NM' ]
print('models:',models)
for model in models:
print('model:',model)
mic = nuda.matter.setupMicro( model = model )
models: ['2013-QMC-NM', '2016-MBPT-AM', '2016-QMC-NM', '2020-MBPT-AM', '2024-QMC-NM']
model: 2013-QMC-NM
model -> 2013-QMC-NM
model: 2016-MBPT-AM
model -> 2016-MBPT-AM
model: 2016-QMC-NM
model -> 2016-QMC-NM
model: 2020-MBPT-AM
model -> 2020-MBPT-AM
model: 2024-QMC-NM
model -> 2024-QMC-NM
plot:
nuda.fig.matter_setupMicro_err_NM_fig( None, models )
Plot name: None
model -> 2013-QMC-NM
model: 2013-QMC-NM
model -> 2016-MBPT-AM
model: 2016-MBPT-AM
model -> 2016-QMC-NM
model: 2016-QMC-NM
model -> 2020-MBPT-AM
model: 2020-MBPT-AM
model -> 2024-QMC-NM
model: 2024-QMC-NM
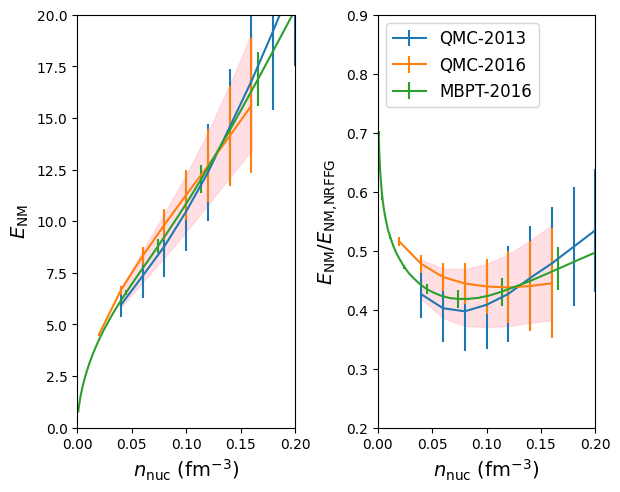
We now define a reference band in NM:
We first choose as set of calculations existing in nuda’s library. We first need to know what are the calculations available:
models, models_lower = nuda.matter.micro_models()
print('models:',models)
models: ['1981-VAR-AM-FP', '1998-VAR-AM-APR', '1998-VAR-AM-APR-fit', '2012-AFDMC-NM-RES-1', '2012-AFDMC-NM-RES-2', '2012-AFDMC-NM-RES-3', '2012-AFDMC-NM-RES-4', '2012-AFDMC-NM-RES-5', '2012-AFDMC-NM-RES-6', '2012-AFDMC-NM-RES-7', '2012-AFDMC-NM-FIT-1', '2012-AFDMC-NM-FIT-2', '2012-AFDMC-NM-FIT-3', '2012-AFDMC-NM-FIT-4', '2012-AFDMC-NM-FIT-5', '2012-AFDMC-NM-FIT-6', '2012-AFDMC-NM-FIT-7', '2022-AFDMC-NM', '2024-BHF-AM-2BF-Av8p', '2024-BHF-AM-2BF-Av18', '2024-BHF-AM-2BF-BONN', '2024-BHF-AM-2BF-CDBONN', '2024-BHF-AM-2BF-NSC97a', '2024-BHF-AM-2BF-NSC97b', '2024-BHF-AM-2BF-NSC97c', '2024-BHF-AM-2BF-NSC97d', '2024-BHF-AM-2BF-NSC97e', '2024-BHF-AM-2BF-NSC97f', '2024-BHF-AM-2BF-SSCV14', '2024-BHF-AM-23BF-Av8p', '2024-BHF-AM-23BF-Av18', '2024-BHF-AM-23BF-BONN', '2024-BHF-AM-23BF-CDBONN', '2024-BHF-AM-23BF-NSC97a', '2024-BHF-AM-23BF-NSC97b', '2024-BHF-AM-23BF-NSC97c', '2024-BHF-AM-23BF-NSC97d', '2024-BHF-AM-23BF-NSC97e', '2024-BHF-AM-23BF-NSC97f', '2024-BHF-AM-23BF-SSCV14', '2008-QMC-NM-swave', '2010-QMC-NM-AV4', '2009-DLQMC-NM', '2013-QMC-NM', '2014-AFQMC-NM', '2016-QMC-NM', '2016-MBPT-AM', '2018-QMC-NM', '2024-QMC-NM', '2010-MBPT-NM', '2016-MBPT-AM', '2019-MBPT-AM-L59', '2019-MBPT-AM-L69', '2020-MBPT-AM', '2024-NLEFT-AM']
Then we select a few of them:
models = [ '2013-QMC-NM', '2016-QMC-NM', '2016-MBPT-AM' ]
The reference band in NM is then calculated in the following way:
band = nuda.matter.setupMicroBand( models, matter='NM' )
band.print_outputs( )
Band in NM
model -> 2013-QMC-NM
model -> 2016-QMC-NM
model -> 2016-MBPT-AM
model -> 2013-QMC-NM
model -> 2016-QMC-NM
model -> 2016-MBPT-AM
model -> 2013-QMC-NM
model -> 2016-QMC-NM
model -> 2016-MBPT-AM
- Print output:
models : ['2013-QMC-NM', '2016-QMC-NM', '2016-MBPT-AM']
den_min: 0.04
den_max: 0.16
den : [0.04 0.052 0.064 0.076 0.088 0.1 0.112 0.124 0.136 0.148 0.16 ]
kfn : [1.06 1.15 1.24 1.31 1.38 1.44 1.49 1.54 1.59 1.64 1.68]
e2a : [ 6.28 7.21 8.1 9. 9.92 10.87 11.85 12.9 13.99 15.11 16.24]
std : [0.46 0.646 0.827 1.007 1.182 1.371 1.594 1.845 2.131 2.461 2.829]
Plot:
den = np.linspace(0.04,0.16,num=7)
print('den:',den)
nuda.fig.matter_setupMicro_band_fig( None, models, den=den, matter='NM' )
den: [0.04 0.06 0.08 0.1 0.12 0.14 0.16]
Plot name: None
model -> 2013-QMC-NM
model: 2013-QMC-NM
model -> 2016-QMC-NM
model: 2016-QMC-NM
model -> 2016-MBPT-AM
model: 2016-MBPT-AM
Band in NM
model -> 2013-QMC-NM
model -> 2016-QMC-NM
model -> 2016-MBPT-AM
model -> 2013-QMC-NM
model -> 2016-QMC-NM
model -> 2016-MBPT-AM