2.5. Pairing gap (micro)#
In this tutorial, you will learn how to employ the pairing gap module.
Import the libraries that will be employed in this tutorial.
# Import numpy
import numpy as np
# Import matplotlib
import matplotlib.pyplot as plt
# Import nucleardatapy package
import nucleardatapy as nuda
You can simply print out the properties of the nuda’s function that we will use:
# Explore the nucleardatapy module to find the correct attribute
print(dir(nuda.matter.setupMicroGap))
['__class__', '__delattr__', '__dict__', '__dir__', '__doc__', '__eq__', '__format__', '__ge__', '__getattribute__', '__getstate__', '__gt__', '__hash__', '__init__', '__init_subclass__', '__le__', '__lt__', '__module__', '__ne__', '__new__', '__reduce__', '__reduce_ex__', '__repr__', '__setattr__', '__sizeof__', '__str__', '__subclasshook__', '__weakref__', 'init_self', 'print_outputs']
Get the full list of models ( matter can be ‘NM’ (default) or ‘SM’ ) :
models, models_lower, models_all, models_all_lower = nuda.matter.micro_gap_models( matter = 'NM' )
print('models:',models)
models: ['2006-BHF-NM-Av18', '2008-BCS-NM', '2008-QMC-NM-swave', '2009-DLQMC-NM', '2010-QMC-NM-AV4', '2017-MBPT-NM-GAP-EMG-450-500-N2LO', '2017-MBPT-NM-GAP-EMG-450-500-N3LO', '2017-MBPT-NM-GAP-EMG-450-700-N2LO', '2017-MBPT-NM-GAP-EMG-450-700-N3LO', '2017-MBPT-NM-GAP-EM-500-N2LO', '2017-MBPT-NM-GAP-EM-500-N3LO', '2022-AFDMC-NM']
Fix one model for which we want to extract the data:
model = '2006-BHF-NM'
Instantiate the object gap with the pairing gap data:
pheno = nuda.matter.setupMicroGap( model = model )
pheno.print_outputs()
-> model: 2006-BHF-NM
setup_micro_gap: The model name 2006-BHF-NM is not in the list of models.
setup_micro_gap: list of models: ['2006-BHF-NM-Av18', '2008-BCS-NM', '2008-QMC-NM-swave', '2009-DLQMC-NM', '2010-QMC-NM-AV4', '2017-MBPT-NM-GAP-EMG-450-500-N2LO', '2017-MBPT-NM-GAP-EMG-450-500-N3LO', '2017-MBPT-NM-GAP-EMG-450-700-N2LO', '2017-MBPT-NM-GAP-EMG-450-700-N3LO', '2017-MBPT-NM-GAP-EM-500-N2LO', '2017-MBPT-NM-GAP-EM-500-N3LO', '2022-AFDMC-NM']
setup_micro_gap: -- Exit the code --
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[5], line 1
----> 1 pheno = nuda.matter.setupMicroGap( model = model )
2 pheno.print_outputs()
File ~/work/nucleardatapy/nucleardatapy/version-1.0/nucleardatapy/matter/setup_micro_gap.py:91, in setupMicroGap.__init__(self, model, matter)
89 print('setup_micro_gap: list of models:',models)
90 print('setup_micro_gap: -- Exit the code --')
---> 91 exit()
92 #
93 if model.lower() == '2006-bhf-nm-av18':
94 #
NameError: name 'exit' is not defined
Fix a list of params instead of a unique one
models, models_lower, models_all, models_all_lower = nuda.matter.micro_gap_models( matter = 'NM' )
print('models:',models)
for model in models:
print('model:',model)
gap = nuda.matter.setupMicroGap( model = model )
models: ['2006-BHF-NM', '2008-BCS-NM', '2008-QMC-NM-swave', '2009-DLQMC-NM', '2010-QMC-NM-AV4', '2017-MBPT-NM-GAP-EMG-450-500-N2LO', '2017-MBPT-NM-GAP-EMG-450-500-N3LO', '2017-MBPT-NM-GAP-EMG-450-700-N2LO', '2017-MBPT-NM-GAP-EMG-450-700-N3LO', '2017-MBPT-NM-GAP-EM-500-N2LO', '2017-MBPT-NM-GAP-EM-500-N3LO', '2022-AFDMC-NM']
model: 2006-BHF-NM
-> model: 2006-BHF-NM
model: 2008-BCS-NM
-> model: 2008-BCS-NM
model: 2008-QMC-NM-swave
-> model: 2008-QMC-NM-swave
model: 2009-DLQMC-NM
-> model: 2009-DLQMC-NM
model: 2010-QMC-NM-AV4
-> model: 2010-QMC-NM-AV4
model: 2017-MBPT-NM-GAP-EMG-450-500-N2LO
-> model: 2017-MBPT-NM-GAP-EMG-450-500-N2LO
model: 2017-MBPT-NM-GAP-EMG-450-500-N3LO
-> model: 2017-MBPT-NM-GAP-EMG-450-500-N3LO
model: 2017-MBPT-NM-GAP-EMG-450-700-N2LO
-> model: 2017-MBPT-NM-GAP-EMG-450-700-N2LO
model: 2017-MBPT-NM-GAP-EMG-450-700-N3LO
-> model: 2017-MBPT-NM-GAP-EMG-450-700-N3LO
model: 2017-MBPT-NM-GAP-EM-500-N2LO
-> model: 2017-MBPT-NM-GAP-EM-500-N2LO
model: 2017-MBPT-NM-GAP-EM-500-N3LO
-> model: 2017-MBPT-NM-GAP-EM-500-N3LO
model: 2022-AFDMC-NM
-> model: 2022-AFDMC-NM
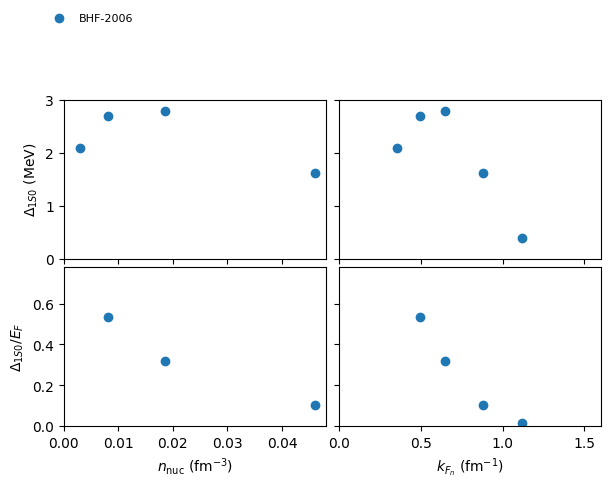
plot 1S0 pairing gap in NM:
nuda.fig.matter_setupMicro_gap_1s0_fig( None, models, matter = 'NM' )
Plot name: None
models: ['2006-BHF-NM', '2008-BCS-NM', '2008-QMC-NM-swave', '2009-DLQMC-NM', '2010-QMC-NM-AV4', '2017-MBPT-NM-GAP-EMG-450-500-N2LO', '2017-MBPT-NM-GAP-EMG-450-500-N3LO', '2017-MBPT-NM-GAP-EMG-450-700-N2LO', '2017-MBPT-NM-GAP-EMG-450-700-N3LO', '2017-MBPT-NM-GAP-EM-500-N2LO', '2017-MBPT-NM-GAP-EM-500-N3LO', '2022-AFDMC-NM']
-> model: 2006-BHF-NM
-> model: 2008-BCS-NM
-> model: 2008-QMC-NM-swave
-> model: 2009-DLQMC-NM
-> model: 2010-QMC-NM-AV4
-> model: 2017-MBPT-NM-GAP-EMG-450-500-N2LO
-> model: 2017-MBPT-NM-GAP-EMG-450-500-N3LO
-> model: 2017-MBPT-NM-GAP-EMG-450-700-N2LO
-> model: 2017-MBPT-NM-GAP-EMG-450-700-N3LO
-> model: 2017-MBPT-NM-GAP-EM-500-N2LO
-> model: 2017-MBPT-NM-GAP-EM-500-N3LO
-> model: 2022-AFDMC-NM
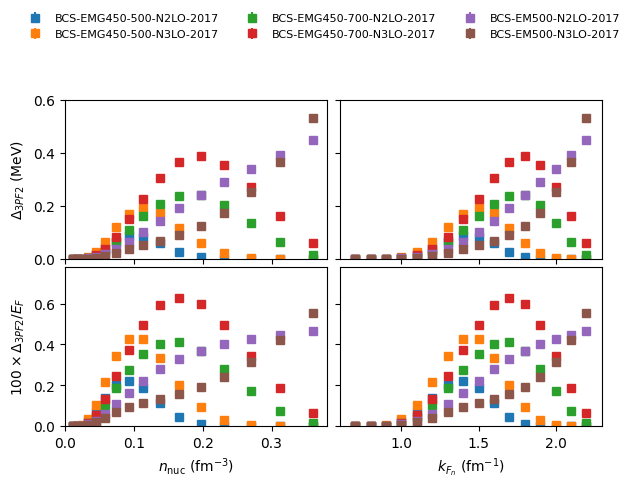
plot 3PGF2 pairing gap in NM:
nuda.fig.matter_setupMicro_gap_3pf2_fig( None, models, matter = 'NM' )
Plot name: None
models: ['2006-BHF-NM', '2008-BCS-NM', '2008-QMC-NM-swave', '2009-DLQMC-NM', '2010-QMC-NM-AV4', '2017-MBPT-NM-GAP-EMG-450-500-N2LO', '2017-MBPT-NM-GAP-EMG-450-500-N3LO', '2017-MBPT-NM-GAP-EMG-450-700-N2LO', '2017-MBPT-NM-GAP-EMG-450-700-N3LO', '2017-MBPT-NM-GAP-EM-500-N2LO', '2017-MBPT-NM-GAP-EM-500-N3LO', '2022-AFDMC-NM']
-> model: 2006-BHF-NM
-> model: 2008-BCS-NM
-> model: 2008-QMC-NM-swave
-> model: 2009-DLQMC-NM
-> model: 2010-QMC-NM-AV4
-> model: 2017-MBPT-NM-GAP-EMG-450-500-N2LO
-> model: 2017-MBPT-NM-GAP-EMG-450-500-N3LO
-> model: 2017-MBPT-NM-GAP-EMG-450-700-N2LO
-> model: 2017-MBPT-NM-GAP-EMG-450-700-N3LO
-> model: 2017-MBPT-NM-GAP-EM-500-N2LO
-> model: 2017-MBPT-NM-GAP-EM-500-N3LO
-> model: 2022-AFDMC-NM
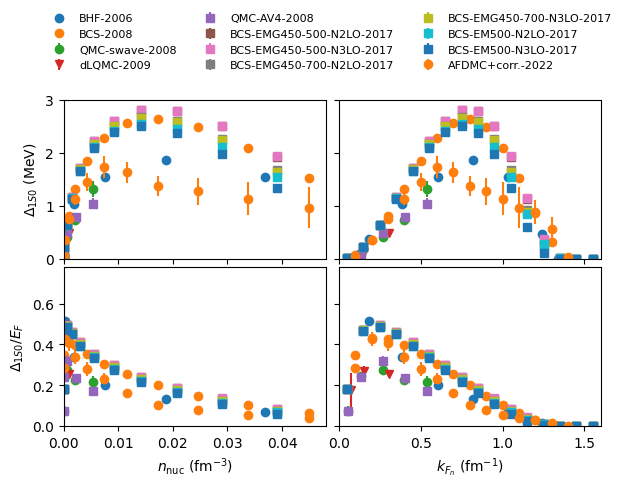
plot 1S0 pairing gap in SM:
models, models_lower, models_all, models_all_lower = nuda.matter.micro_gap_models( matter = 'SM' )
print('models:',models)
nuda.fig.matter_setupMicro_gap_1s0_fig( None, models, matter = 'SM' )
models: ['2006-BHF-SM']
Plot name: None
models: ['2006-BHF-SM']
-> model: 2006-BHF-SM