4.2. Two-neutron and two-proton separation energies#
In this tutorial, you will learn how to obtain the two-neutron and two-proton separation energies from the experimental nuclear chart.
Import the libraries that will be employed in this tutorial.
# Import numpy
import numpy as np
# Import matplotlib
import matplotlib.pyplot as plt
# Import nucleardatapy package
import nucleardatapy as nuda
You can simply print out the properties of the nuda’s function that we will use:
# Explore the nucleardatapy module to find the correct attribute
print(dir(nuda.nuc.setupBEExp))
['D3n', 'D3p', 'S2n', 'S2p', '__class__', '__delattr__', '__dict__', '__dir__', '__doc__', '__eq__', '__format__', '__ge__', '__getattribute__', '__getstate__', '__gt__', '__hash__', '__init__', '__init_subclass__', '__le__', '__lt__', '__module__', '__ne__', '__new__', '__reduce__', '__reduce_ex__', '__repr__', '__setattr__', '__sizeof__', '__str__', '__subclasshook__', '__weakref__', 'isotones', 'isotopes', 'print_outputs', 'select', 'select_year']
Get the full list of tables:
tables, tables_lower = nuda.nuc.be_exp_tables()
print('tables:',tables)
tables: ['AME']
Get the versions associated to this table:
versions, versions_lower = nuda.nuc.be_exp_versions(table = 'AME')
print('versions:',versions)
versions: ['2020', '2016', '2012']
Extract data from a given table and version:
mas = nuda.nuc.setupBEExp( table = 'AME', version = '2020' )
print('number of nuclei:',mas.nbNuc)
Oldest discovery is from: 1896
Most recent discovery is from: 2020
dist: [ 20. 19. 118. 170. 253. 620. 510. 888. 632. 394. 310. 252.
72. 1569. 0. 0. 0. 0. 0. 0.]
number of nuclei: 5827
Compute the two-neutron separation energies:
Zref = 50
s2n = mas.S2n( Zmin = Zref, Zmax = Zref )
---------------------------------------------------------------------------
TypeError Traceback (most recent call last)
Cell In[6], line 2
1 Zref = 50
----> 2 s2n = mas.S2n( Zmin = Zref, Zmax = Zref )
TypeError: setupBEExp.S2n() got an unexpected keyword argument 'Zmin'
Compute the two-proton separation energies:
Nref = 50
s2p = mas.S2p( Nmin = Nref, Nmax = Nref )
print results:
print('N(Z=50):',s2n.S2n_N)
print('S2n(Z=50):',s2n.S2n)
print()
print('Z(N=50):',s2p.S2p_Z)
print('S2p(N=50):',s2p.S2p)
N(Z=50): [52 54 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77
78 79 80 81 82 83 84 85]
S2n(Z=50): [23.9226342 22.8396342 21.8696342 21.3166342 20.8586342 20.2606342
19.9146342 19.4516342 18.9556842 18.5317342 18.0473192 17.8483802
17.1088792 16.5064882 16.2694542 15.8099342 15.5875342 15.2742342
14.9849342 14.7607342 14.4341342 14.2216342 13.9261342 13.7189342
13.4886342 13.2636342 12.9138342 12.8166342 12.5570342 9.7515342
6.0300342 5.9007342]
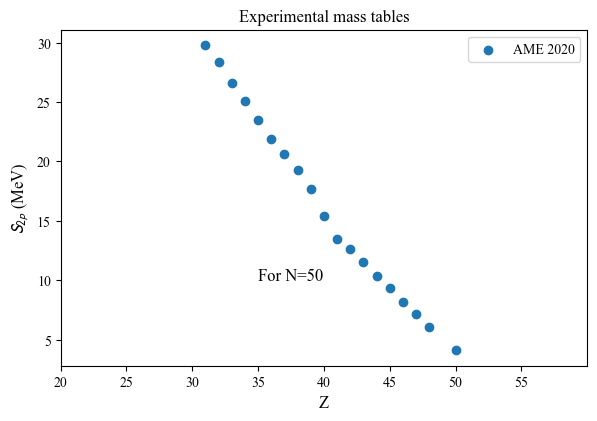
Z(N=50): [31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 50]
S2p(N=50): [29.7959412 28.3444412 26.6192412 25.1105412 23.4836412 21.8959172
20.6007432 19.2338942 17.6913392 15.4288622 13.5047412 12.6139812
11.5460412 10.3533512 9.3128412 8.1769412 7.1409412 6.0349412
4.0879412]
Plot:
nuda.fig.nuc_setupBEExp_S2n_fig( None, tables, versions, Zref = 50 )
Plot name: None
Tables: ['AME']
Zref: 50
Oldest discovery is from: 1896
Most recent discovery is from: 2020
dist: [ 20. 19. 118. 170. 253. 620. 510. 888. 632. 394. 310. 252.
72. 1569. 0. 0. 0. 0. 0. 0.]
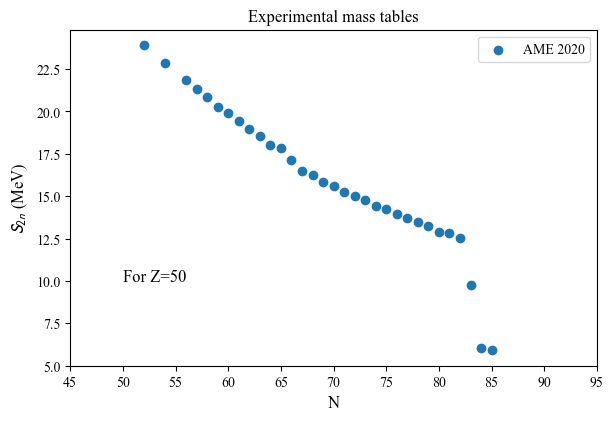
nuda.fig.nuc_setupBEExp_S2p_fig( None, tables, versions, Nref = 50 )
Plot name: None
Tables: ['AME']
Nref: 50
Plot name: None
Oldest discovery is from: 1896
Most recent discovery is from: 2020
dist: [ 20. 19. 118. 170. 253. 620. 510. 888. 632. 394. 310. 252.
72. 1569. 0. 0. 0. 0. 0. 0.]