4.3. Experimental odd-even mass staggering (OEMS)#
In this tutorial, you will learn how to calculate the OEMS from the experimental nuclear chart (using the 3-point formula).
Import the libraries that will be employed in this tutorial.
# Import numpy
import numpy as np
# Import matplotlib
import matplotlib.pyplot as plt
# Import nucleardatapy package
import nucleardatapy as nuda
You can simply print out the properties of the nuda’s function that we will use:
# Explore the nucleardatapy module to find the correct attribute
print(dir(nuda.nuc.setupBEExp))
['D3n', 'D3p', 'S2n', 'S2p', '__class__', '__delattr__', '__dict__', '__dir__', '__doc__', '__eq__', '__format__', '__ge__', '__getattribute__', '__getstate__', '__gt__', '__hash__', '__init__', '__init_subclass__', '__le__', '__lt__', '__module__', '__ne__', '__new__', '__reduce__', '__reduce_ex__', '__repr__', '__setattr__', '__sizeof__', '__str__', '__subclasshook__', '__weakref__', 'isotones', 'isotopes', 'print_outputs', 'select', 'select_year']
Get the full list of tables:
tables, tables_lower = nuda.nuc.be_exp_tables()
print('tables:',tables)
tables: ['AME']
Get the versions associated to this table:
versions, versions_lower = nuda.nuc.be_exp_versions(table = 'AME')
print('versions:',versions)
versions: ['2020', '2016', '2012']
Extract data from a given table and version:
mas = nuda.nuc.setupBEExp( table = 'AME', version = '2020' )
print('number of nuclei:',mas.nbNuc)
Oldest discovery is from: 1896
Most recent discovery is from: 2020
dist: [ 20. 19. 118. 170. 253. 620. 510. 888. 632. 394. 310. 252.
72. 1569. 0. 0. 0. 0. 0. 0.]
number of nuclei: 5827
Compute the neutron EOMS:
Zref = 50
d3n = mas.D3n( Zmin = Zref, Zmax = Zref )
---------------------------------------------------------------------------
TypeError Traceback (most recent call last)
Cell In[6], line 2
1 Zref = 50
----> 2 d3n = mas.D3n( Zmin = Zref, Zmax = Zref )
TypeError: setupBEExp.D3n() got an unexpected keyword argument 'Zmin'
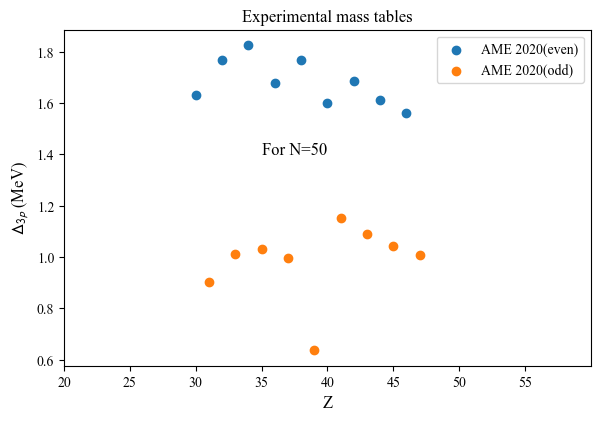
Compute the proton EOMS:
Nref = 50
d3p = mas.D3p( Nmin = Nref, Nmax = Nref )
print results:
print('N_even(Z=50):',d3n.D3n_N_even)
print('D3n(Z=50):',d3n.D3n_even)
print()
print('Z_even(N=50):',d3p.D3p_Z_even)
print('D3p(N=50):',d3p.D3p_even)
N_even(Z=50): [56 58 60 62 64 66 68 70 72 74 76 78 80 82 84]
D3n(Z=50): [1.429 1.499 1.5575 1.5215 1.378762 1.310207 1.42145 1.4669
1.43435 1.3773 1.33315 1.3305 1.2042 2.47715 0.68105 ]
Z_even(N=50): [30 32 34 36 38 40 42 44 46]
D3p(N=50): [1.6296 1.76645 1.82555 1.679275 1.767128 1.59795 1.68654 1.61045
1.5605 ]
Plot nuclear chart:
nuda.fig.nuc_setupBEExp_D3n_fig( None, tables, versions, Zref = 50 )
Plot name: None
Tables: ['AME']
Zref: 50
Oldest discovery is from: 1896
Most recent discovery is from: 2020
dist: [ 20. 19. 118. 170. 253. 620. 510. 888. 632. 394. 310. 252.
72. 1569. 0. 0. 0. 0. 0. 0.]
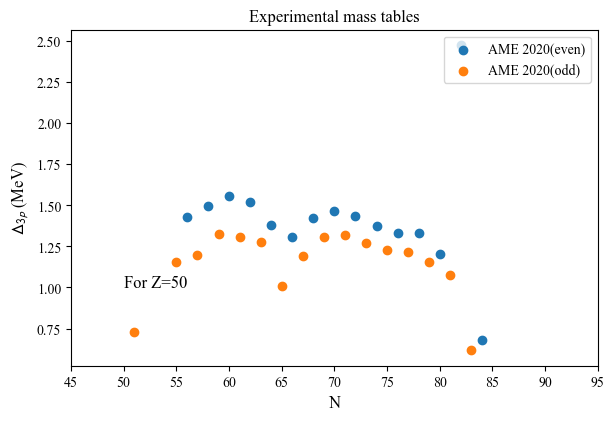
nuda.fig.nuc_setupBEExp_D3p_fig( None, tables, versions, Nref = 50 )
Plot name: None
Tables: ['AME']
Nref: 50
Oldest discovery is from: 1896
Most recent discovery is from: 2020
dist: [ 20. 19. 118. 170. 253. 620. 510. 888. 632. 394. 310. 252.
72. 1569. 0. 0. 0. 0. 0. 0.]